EX2: CBC-GW Injection#
In this example we load an LVK posterior from zenodo and compute the FI evidence for the CBC model.
% load_ext autoreload
% autoreload 2
% matplotlib inline
UsageError: Line magic function `%` not found.
import bilby
import numpy as np
import os, shutil
from collections import namedtuple
import logging
import matplotlib.pyplot as plt
from funnel.plotting import plot_fi_evidence_results
from funnel.fi_core import get_fi_lnz_list
np.random.seed(42)
CLEAN = False
OUTDIR = 'out_gwinj'
if os.path.exists(OUTDIR) and CLEAN:
shutil.rmtree(OUTDIR)
os.makedirs(OUTDIR, exist_ok=True)
Simple 2-parameter inferece#
Nested sampling LnZ Calculation#
bilby_logger = logging.getLogger("bilby")
bilby_logger.setLevel(logging.CRITICAL)
label = "injection"
res_fname = f"{OUTDIR}/{label}_result.json"
if not os.path.exists(res_fname):
# Simulate signal
duration, sampling_freq, min_freq = 4, 1024., 20
injection_parameters = dict(
mass_1=36.0, mass_2=29.0, # 2 mass parameters
a_1=0.1, a_2=0.1, tilt_1=0.0, tilt_2=0.0, phi_12=0.0, phi_jl=0.0, # 6 spin parameters
ra=1.375, dec=-1.2108, luminosity_distance=2000.0, theta_jn=0.0, # 7 extrinsic parameters
psi=2.659, phase=1.3,
geocent_time=1126259642.413,
)
inj_m1, inj_m2 = injection_parameters['mass_1'], injection_parameters['mass_2']
inj_chirp_mass = bilby.gw.conversion.component_masses_to_chirp_mass(inj_m1, inj_m2)
inj_q = bilby.gw.conversion.component_masses_to_mass_ratio(inj_m1, inj_m2)
waveform_generator = bilby.gw.WaveformGenerator(
duration=duration,
sampling_frequency=sampling_freq,
frequency_domain_source_model=bilby.gw.source.lal_binary_black_hole,
parameter_conversion=bilby.gw.conversion.convert_to_lal_binary_black_hole_parameters,
waveform_arguments=dict(
waveform_approximant="IMRPhenomD",
reference_frequency=20.0,
minimum_frequency=min_freq,
)
)
# Inject the signal into 1 detectors LIGO-Hanford (H1) at design sensitivity
ifos = bilby.gw.detector.InterferometerList(["H1"])
ifos.set_strain_data_from_power_spectral_densities(
sampling_frequency=sampling_freq,
duration=duration,
start_time=injection_parameters["geocent_time"] - 2,
)
ifos.inject_signal(
waveform_generator=waveform_generator, parameters=injection_parameters
)
# We sample in chirp-mass and mass-ratio, however--these are quite un-astrophysical priors
# but in post-processing convert to uniform-in-component masses
priors = bilby.gw.prior.BBHPriorDict()
for key in [
"a_1",
"a_2",
"tilt_1",
"tilt_2",
"phi_12",
"phi_jl",
"psi",
"ra",
"dec",
"geocent_time",
"phase",
"theta_jn",
"luminosity_distance",
]:
priors[key] = injection_parameters[key]
priors["chirp_mass"] = bilby.gw.prior.UniformInComponentsChirpMass(
minimum=inj_chirp_mass - 5,
maximum=inj_chirp_mass + 5
)
# Perform a check that the prior does not extend to a parameter space longer than the data
priors.validate_prior(duration, min_freq);
likelihood = bilby.gw.GravitationalWaveTransient(
interferometers=ifos,
waveform_generator=waveform_generator,
)
result = bilby.run_sampler(
likelihood=likelihood,
priors=priors,
sampler="dynesty",
npoints=1000,
dlogz=0.1,
injection_parameters=injection_parameters,
outdir=OUTDIR,
label=label,
conversion_function=bilby.gw.conversion.generate_all_bbh_parameters,
result_class=bilby.gw.result.CBCResult,
)
else:
result = bilby.gw.result.CBCResult.from_json(filename=res_fname)
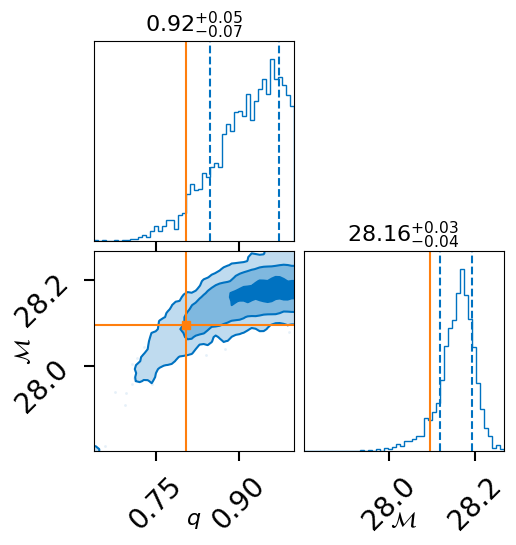
fig = result.plot_corner()
post = result.posterior[['mass_ratio', 'chirp_mass', 'log_prior', 'log_likelihood']]
lnz = result.log_evidence
lnz_err = result.log_evidence_err
fig
print(f"lnZ = {lnz:.2f} +/- {lnz_err:.2f}")
print(f"lnBF = {result.log_bayes_factor:.2f}")
lnZ = -1984.29 +/- 0.10
lnBF = 70.70
NOTE: the LVK stores the log-likelihood - noise log-likelihood in the column labeled ‘log-likelihood’. So, this time instead of computing the FI LnZ, we will compute the FI LnBF.
Compute FI evidence#
lnz_file = f"{OUTDIR}/lnz.npz"
lnzs, r_vals, _ = get_fi_lnz_list(post, r_vals=np.geomspace(1e1, 2e5, 1000), num_ref_params=100, cache_fn=lnz_file)
|funnel|INFO| Calculating FI LnZ with 100 reference points and a posterior of size: (2653, 2)
plt_kwgs = dict(lnzs=lnzs, r_vals=r_vals, sampling_lnz=[result.log_bayes_factor])
fig = plot_fi_evidence_results(**plt_kwgs)
fig.tight_layout()
fig.gca().set_ylabel(r"ln BF");
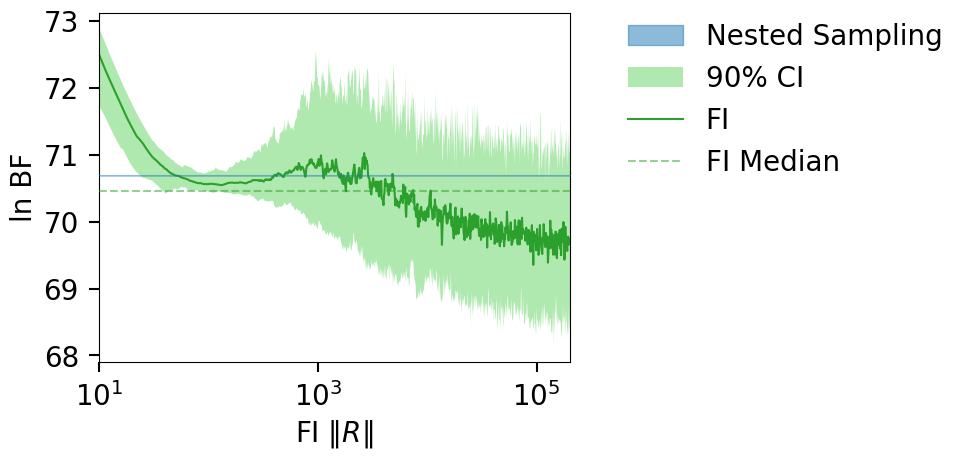
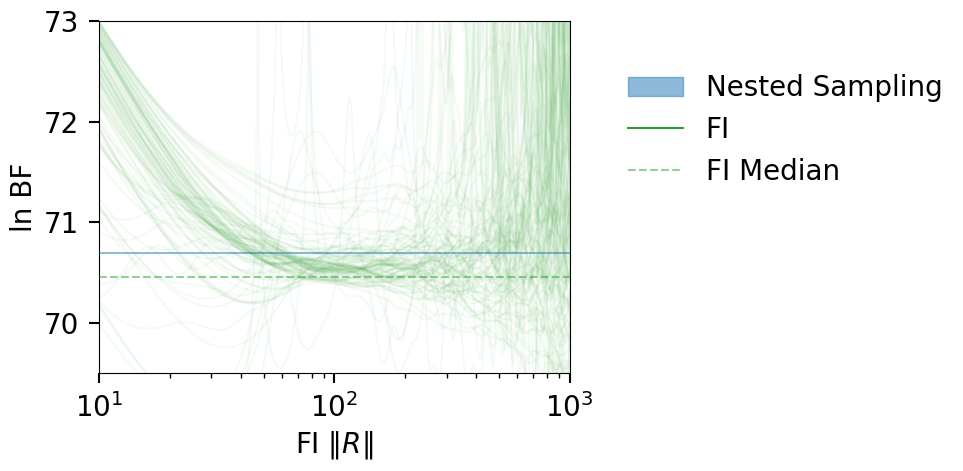
fig = plot_fi_evidence_results(**plt_kwgs, plot_all_lnzs=True, plot_ci=False)
fig.tight_layout()
ax = fig.gca()
ax.set_ylabel(r"ln BF");
ax.set_xlim(1e1, 1e3);
ax.set_ylim(69.5, 73)
(69.5, 73.0)
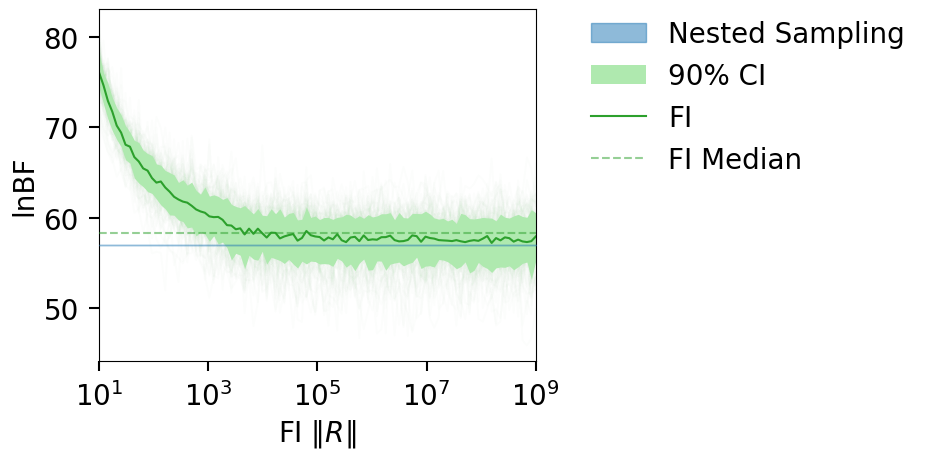
15-D parameter inference#
We download a pre-run LVK injection and run the FI LnZ computation on it. Link to result: https://zenodo.org/record/7884973
import bilby
# load the LVK posterior
FPATH = '128_cpus_v1_0_result.hdf5'
LVK_data = namedtuple("LVK_data", "posterior, lnz, lnz_err, lnBF")
def load_lvk_data(fpath):
r = bilby.gw.result.CBCResult.from_hdf5(fpath)
post = r.posterior
sampling_params = r.search_parameter_keys + ['log_likelihood', 'log_prior', 'luminosity_distance']
lnz, lnz_err = r.log_evidence, r.log_evidence_err
lnBF = r.log_bayes_factor
post = post[sampling_params]
post = post.loc[:, post.nunique() > 1]
return LVK_data(post, lnz, lnz_err, lnBF)
CBC_data = load_lvk_data(FPATH)
print(f"Posterior shape: {CBC_data.posterior.shape}")
print(f"LnZ: {CBC_data.lnz:.2f} +/- {CBC_data.lnz_err:.2f}")
print(f"LnBF: {CBC_data.lnBF}")
CBC_data.posterior.head().T
Posterior shape: (15196, 17)
LnZ: -16332.68 +/- 0.11
LnBF: 56.96249503300896
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| chirp_mass | 17.303518 | 17.137896 | 17.406246 | 17.221114 | 17.224999 |
| mass_ratio | 0.640732 | 0.363822 | 0.475442 | 0.716670 | 0.535031 |
| a_1 | 0.122786 | 0.126647 | 0.208230 | 0.124358 | 0.064088 |
| a_2 | 0.294212 | 0.671431 | 0.858286 | 0.436716 | 0.239654 |
| tilt_1 | 2.671935 | 1.089454 | 0.899129 | 2.487101 | 1.332991 |
| tilt_2 | 0.617993 | 1.095211 | 1.673787 | 1.440678 | 1.530625 |
| phi_12 | 5.277061 | 5.083531 | 0.924704 | 4.632946 | 5.622373 |
| phi_jl | 5.421349 | 0.331844 | 3.584094 | 4.388201 | 4.657011 |
| cos_theta_jn | 0.649265 | -0.750138 | -0.906039 | -0.887552 | -0.956641 |
| psi | 3.033949 | 1.530611 | 2.617934 | 1.037721 | 3.061500 |
| phase | 3.838546 | 4.858204 | 0.359604 | 1.671526 | 3.573420 |
| azimuth | 3.105585 | 2.906103 | 2.371659 | 2.250562 | 1.480921 |
| zenith | 0.552605 | 0.251147 | 0.141375 | 0.281922 | 0.242988 |
| time_jitter | -0.000104 | 0.000073 | -0.000216 | 0.000176 | 0.000130 |
| log_likelihood | 59.964813 | 63.408287 | 63.456029 | 63.735976 | 63.985682 |
| log_prior | -7.725553 | -6.777387 | -7.630868 | -7.647782 | -7.011267 |
| luminosity_distance | 835.104547 | 644.254930 | 1117.045547 | 1151.937210 | 1433.822042 |
NOTE: the log-likelihood column is actually the lnl-noise Lnl (the log-likelihood-ratio).
Compute FI LnZ#
def run_fi_computer(CBC_data, frac_samp=0.1, n_ref_points=1000, outdir='out_inj_downsampled', clean=False):
if os.path.exists(outdir) and clean:
shutil.rmtree(outdir)
os.makedirs(outdir, exist_ok=True)
# USING DOWNSAMPLED POSTERIOR SAMPLES
n_total = len(CBC_data.posterior)
n_samp = int(n_total * frac_samp)
post = CBC_data.posterior.sample(
n_samp,
weights=np.exp(CBC_data.posterior.log_likelihood)
)
print(f"Using {100 * frac_samp:.2f}% of posterior, and {n_ref_points} reference points.")
lnzs, r_vals, _ = get_fi_lnz_list(
post,
num_ref_params=n_ref_points,
r_vals=np.geomspace(10, 1e9, 100),
cache_fn=f'{outdir}/lnzs.npz',
weight_samples_by_lnl=True,
)
return lnzs, r_vals
Using a downsampled posterior#
OUTDIR = 'out_inj_downsampled'
CLEAN = False
lnzs, r_vals = run_fi_computer(CBC_data, frac_samp=0.1, n_ref_points=1000, outdir=OUTDIR, clean=CLEAN)
Using 10.00% of posterior, and 1000 reference points.
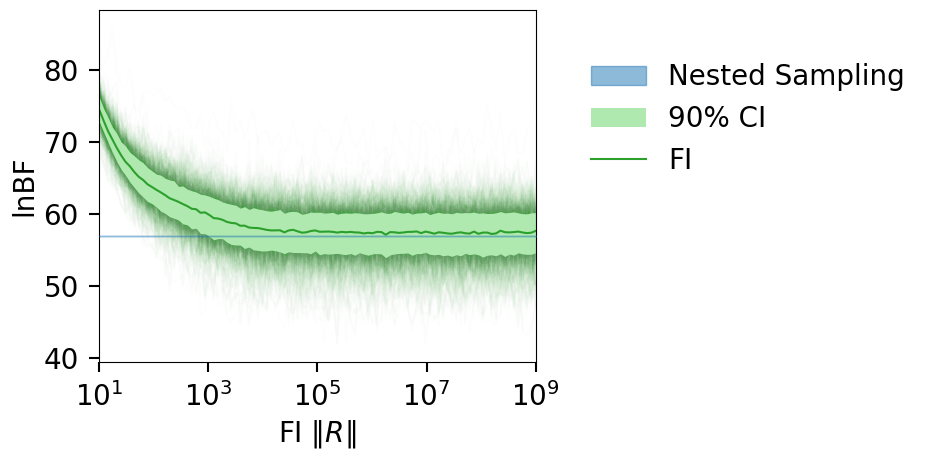
plt_kwgs = dict(lnzs=lnzs, r_vals=r_vals, sampling_lnz=[CBC_data.lnBF])
fig = plot_fi_evidence_results(**plt_kwgs, plot_all_lnzs=True, plt_kwgs=dict(alpha=0.01))
fig.gca().set_ylabel("lnBF");
def histogram_fi_lnzs(lnzs, rvals, rval_threshold, true=None):
idx = np.argmin(np.abs(rvals - rval_threshold))
lnz = lnzs[:, idx:].ravel()
print(f"%samples above {rval_threshold}: {len(lnz)/len(lnzs.ravel()) * 100:.2f}")
fig, ax = plt.subplots(1, 1, figsize=(5, 4))
ax.hist(lnz, bins=100, density=True, histtype='step', lw=2, color='tab:green')
if true is not None:
ax.axvline(true, ls='--', color='tab:blue', lw=2)
ax.set_xlabel(r"FI lnBF")
ax.set_ylabel("pdf")
return fig
fig = histogram_fi_lnzs(lnzs, r_vals, rval_threshold=1e7, true=CBC_data.lnBF)
%samples above 10000000.0: 26.00
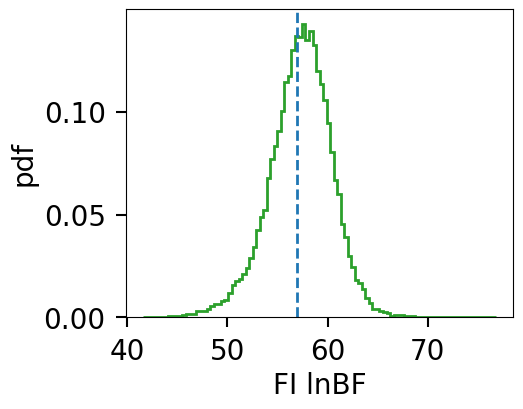
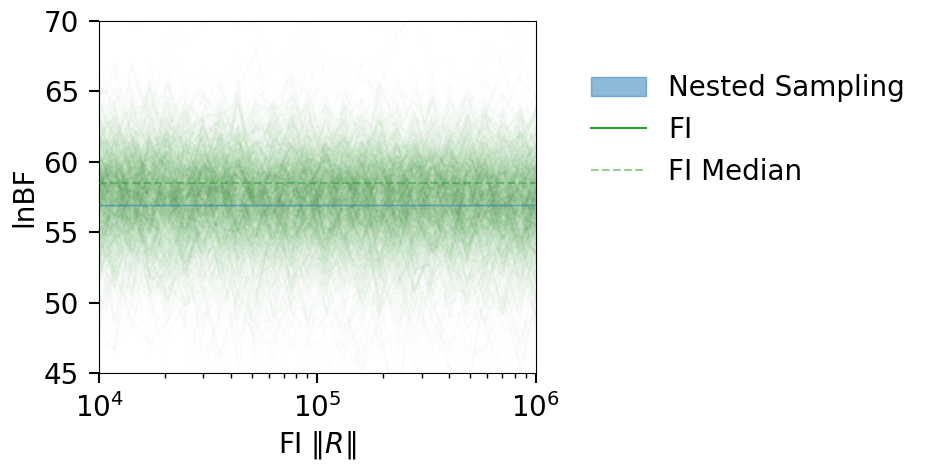
fig = plot_fi_evidence_results(**plt_kwgs, plot_all_lnzs=True, plot_ci=False, plt_kwgs=dict(alpha=0.01))
ax = fig.gca()
ax.set_ylabel("lnBF");
ax.set_xlim(1e4, 1e6);
ax.set_ylim(45, 70);
Using full posterior sample (fewer reference points)#
OUTDIR = 'out_inj2'
CLEAN = False
lnzs, r_vals = run_fi_computer(CBC_data, frac_samp=1, n_ref_points=100, outdir=OUTDIR, clean=CLEAN)
|funnel|INFO| Calculating FI LnZ with 100 reference points and a posterior of size: (15196, 15)
Using 100.00% of posterior, and 1000 reference points.
plt_kwgs = dict(lnzs=lnzs, r_vals=r_vals, sampling_lnz=[CBC_data.lnBF])
fig = plot_fi_evidence_results(**plt_kwgs, plot_all_lnzs=True, plt_kwgs=dict(alpha=0.01))
fig.gca().set_ylabel("lnBF");